Quantized space SVM
The original SVM calculate n^2 pairwise distance between datapoints, and in the case of Kernel SVM, it’s n^2 pairwise kernel distance. Memory consumption is also O(n^2).
It would cause you headache.
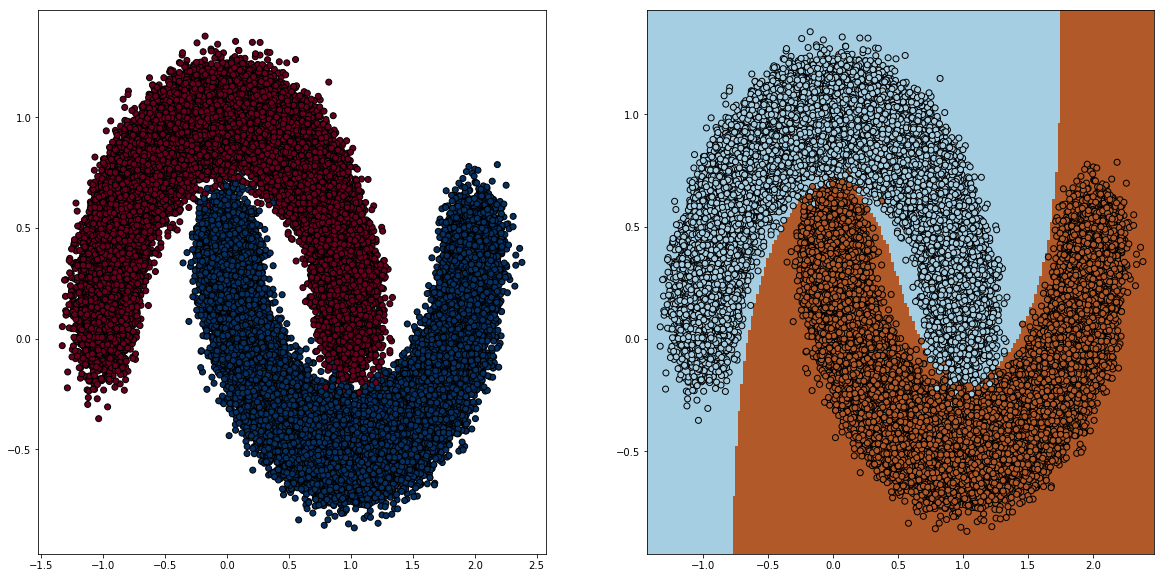
This notebook demonstrate the use of Feature Binning as a way to uniformly subsample the dataset, apply to kernel SVM.
The general idea is to put samples into buckets (bins), and use the bucket or the bucket average as the representative value for all of the samples in that bucket. Number of point falls in to that bin could be used as sample weights if the ML implementation supports.
Number of samples will be reduce drastically, from O(n^2) to worst case O(b*k*t) where b is the number of bins, k is the number of features, and t is the number of classes ( if it’s classification problem).
However, the binned samples density will be altered for sure. But if you care more about the decision boundary more than the actual density, then this is surely helpful.
Now here come the fancy thing: FeatureDiscretizer.
With 100 bins, the sample size reduced from 100k to roughly 5000, using KBinsDiscretizer from Sklearn.
There any some way to obtain the bins, here i choose uniform (All buckets are in the same size, you can choose quantile, where all buckets have the same amount of points). Notice how spread out the samples are.
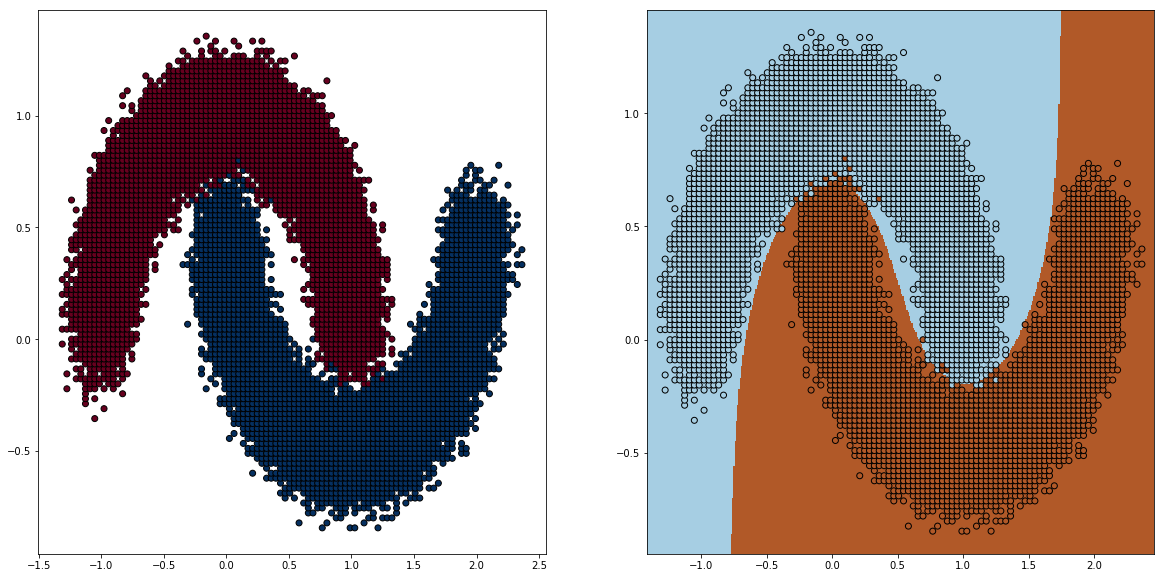
Note that this method doesn’t work well out of the box with density based method like GaussianProcess etc. Density estimations from this method doens’t guaranteed to produce the correct density. I bet there is weighting tricks to make it match :)
Checkout the notebook here.notebook
